COPLA
Using COPLA
COPLA can be used to predict the PTU to which a plasmid belongs. For this just its nucleotide sequence is needed. Draft plasmids are supported by COPLA automatically executing a simple concatenation of their contigs.
If the plasmid ORFeome is uploaded by the user, it will be used by COPLA to obtain a type MOB and MPF according to the user specifications.
Indicating the query topology (linear or circular) is recommended for more accurate MPF typing. If not provided, COPLA uses a circular topology by default. Multifasta input sequences are always processed using a linear topology. For further details see Abby et al. PMID: 26979785.
Finally, if the user provides the taxonomic data asociated with the plasmid host, COPLA will warn the user about changes in the host range of the query's assigned PTU.
How COPLA works
Just as sHSBM, COPLA infers the PTU membership from the similarity relationships between plasmids in the database. After calculating the ANI percentage identity between the query and the reference plasmid set, COPLA inserts the query into the reference network and performs an statistical search for similar plasmids. Finally, the query is assigned to a known PTU, or to a new PTU (labeled as PTU-?) if the algorithm find clues pointing to that outcome. PTUs will not be named for clusters with fewer than 4 members. For the user to evaluate the COPLA PTU assignation of a query plasmid, a score is provided based on the overlap of the graph partitions before and after the query was inserted into the reference network (for additional details see troubleshooting bellow).
COPLA output
COPLA output consists of five different files:
query.ptu_prediction.tsv
File providing the predicted PTU of the query, the graded PTU host range (updated by the taxonomic info of the query if provided by the user), the prediction score, and additional information (see troubleshooting) to help interpret the output.
query.related_plasmids.tsv
List of reference plasmids used for the calculation of the prediction score.
query.ani.tsv
List of ANI percentage identities between the query and reference plasmids. Only plasmids with a detected homology spanning >50% of the shorter genome are reported (Best reciprocal BLASTn hits of 1,000 bp genome fragments with >70% idendity over >70% length. Fragments are obtained using a sliding window algorithm with 200 bp steps). File columns indicate query, reference accession number, ANI, standard deviation of ANI, fragments used for the calculation of ANI and fragments of the reference genome.
query.qry_info.tsv
Other relevant plasmid information: genome length, MOB typing (according to MOBscan), MPF typing (according to CONJScan but with the relaxase HMM profiles updated to those of MOBscan database), replicon typing (based on PlasmidFinder using 80% identity threshold) and AMR genes (based on a BLASTn search against the CARD database)
query.faa
If not provided by the user, this file is the plasmid ORFeome calculated by Prodigal. Prodigal autolearning mode is used for plasmids >100 Kb, smaller plasmids are calculated using Prodigal meta mode.
Troubleshooting: Predicted outcome types
COPLA predictions can result in three different outcomes for PTU membership of the query plasmid. A query could be a member of: (i) a known PTU, (ii) a putative new PTU (provisionally named as "PTU-?"), or (iii) the plasmid remains unclassified (displayed as "-"). The quality of these predictions can be further evaluated with the help of the score output. To assess COPLA performance, we sampled 1,000 RefSeq200 plasmids not present in the reference database (RefSeq84). As shown in Figure 1, results indicate that ~88% of all queries (spanning all three classes) the prediction score a >99%, with the remaining predictions scoring lower values as shown in the figure.
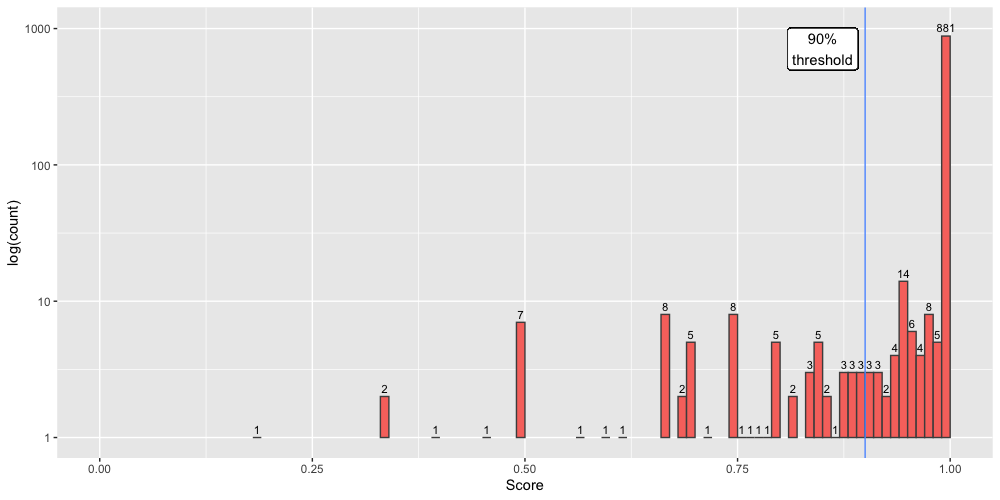
| Figure 1. Score distribution for 1,000 plasmids sampled from RefSeq200, not present in the COPLA reference database (RefSeq84). The figure displays a semilogarithmic plot of the number of plasmids containing each given score. |
To validate the PTU assignment, a score of 90% is recommended. This threshold means that, for a 10-member PTU, the AI has conficting data for clustering 1 of them. As an example, the problematic plasmid could be a cointegrate of plasmids belonging to two different PTUs. Assuming the 90% score threshold, COPLA confidently assigned 93% of the 1,000 samples.
To help understand the different results that COPLA provides, Figure 2 shows several representative schematics.
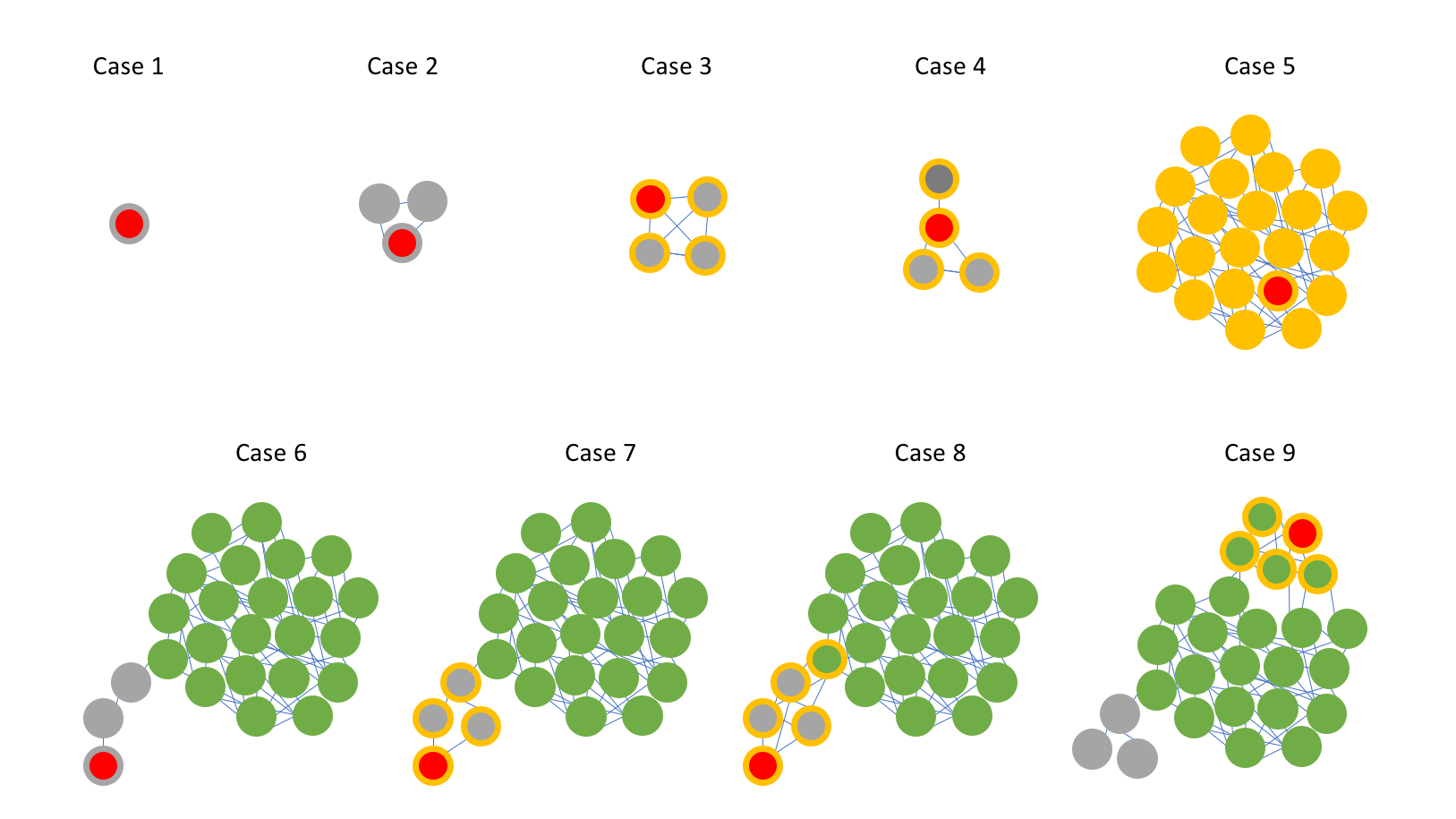
| Figure 2. Representative prediction outcomes. The query plasmid is represented by the node with the red inner circle. For all other nodes, the color of the inner circle represents the PTU assigned in the reference database (i.e. using only RefSeq84 plasmids). The outer rings represent the PTU assigned by COPLA. Yellow represent the PTU assigned to the query, green is a different PTU, and grey colors represent not assigned PTUs. |
When the query is part of a small graph component (4 members or less) the PTU prediction is pretty much trivial (cases 1 to 4):
Case 1 shows a query represented by a singleton, with no ANI relatioships to any reference plasmids, a not uncommon event (355 out of 1,000 samples). In this case, COPLA output includes the sentence: PTU could not be assigned. Query is part of a graph component of size 1.
Case 2 represents a putative PTU, consisting on a set of plasmids unrelated to all other plasmids in the dataset. It is left unnamed as it does not reach the 4-member threshold. In this case (70 out of 1,000 samples) COPLA output includes the sentence: PTU could not be assigned. Query is part of a graph component of size 2|3.
Cases 3 and 4 represent the finding of a new (putative) PTU, by clustering the query with 3 reference plasmids. These reference plasmids could all belong to the same cluster (case 3) or not (with the query acting as an attractor of different plasmids as shown by case 4). These cases (17 out of 1,000 samples) are highlighted in COPLA output by including the sentence: New (putative) PTU. Query is part of a graph component of size 4.
In cases 5 to 7 the query is part of a larger graph component (>4 members), that is, a known PTU. In these cases, the assignation of the query is straightforward. These cases represent the predominant scenarios in which no significant differences in graph partitioning were detected.
In case 5 we have (i) queries for which a PTU can be unequivocally assigned and score is 100% (308 out of 1,000 samples), and (ii) cases in which the score is fewer than 100% (58 out of 1,000 samples with score >=90% and 46 samples with score <90%). COPLA output includes the sentence: Query is a PTU-xxx plasmid.
Case 6 shows a scenario in which no PTU was assigned because of the 4-member threshold. This case (113 out of 1,000 samples) is highlighted in COPLA output by including the sentence: PTU could not be assigned. Query is part of a sHSBM cluster of size 1|2|3.
The finding of a new (putative) PTU is shown in case 7. In this case (8 out of 1,000 samples) COPLA output includes the sentence: New (putative) PTU. Query is part of a sHSBM cluster of size >=4.
Cases 8 and 9 represent scenarios where the inclusion of the query leads to a different partition in the connected component to which the query belongs. Case 8 highlights the case of plasmids (such as cointegrate plasmids) for which the algorithm can not decide unambigously to which PTU should assign the query. Case 9 showcases situations of loosely defined PTUs (showing low intragroup density, such as PTU-FE see ref. [1]). The score provided by COPLA is key to detect and to assess the level of confidence of the PTU prediction.
If the new cluster had fewer than 4 members no PTU is assigned. This case (4 out of 1,000 samples with a score >=90% and 5 with a score <90%) is highlighted in COPLA output by including the sentence: PTU could not be assigned. Query is part of a sHSBM cluster of size 1|2|3.
If the cluster could not be assimilated to a previously known PTU, a new (putative) PTU is defined. In this case (16 out of 1,000 samples, all with score lower than 90%) COPLA output includes the sentence: New (putative) PTU. Query is part of a sHSBM cluster of size >=4.
| [1] Redondo-Salvo, S., Fernandez-Lopez, R., Ruiz., R., Vielva, L., de Toro, M., Rocha, E.P.C., Garcillan-Barcia, M.P., de la Cruz, F. (2020) "Pathways for Horizontal Gene Transfer in Bacteria Revealed by a Global Map of Their Plasmids". Nat Commun 11, 3602 |